New ways to analyze metagenomics data
Are you looking for new tools to analyze your metagenomics data? Are you using MG-RAST, IMG/M or MEGAN for your daily metagenomics work?
JCVI is working on a user friendly alternative that you might be looking for - a new tool kit for metagenomics data visualization and analysis built using the latest web 2.0 technologies.
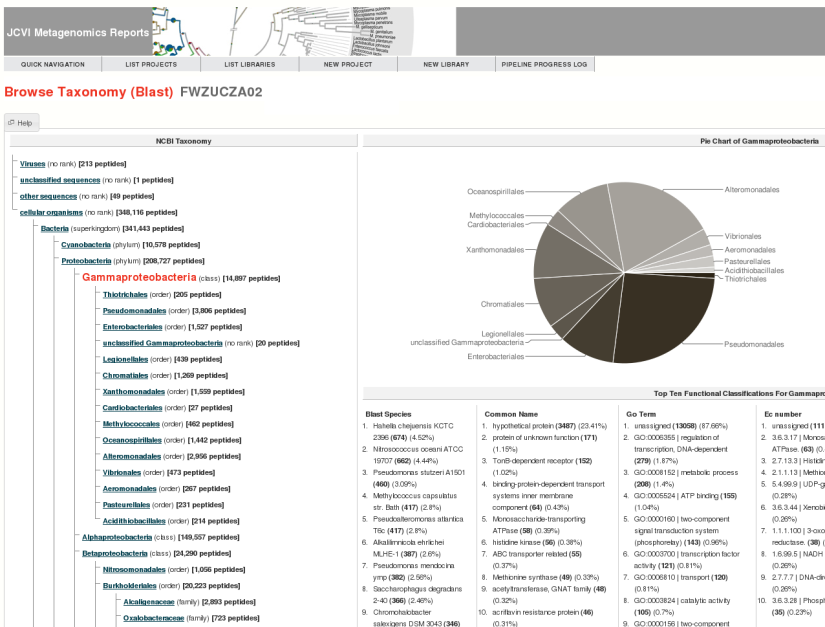
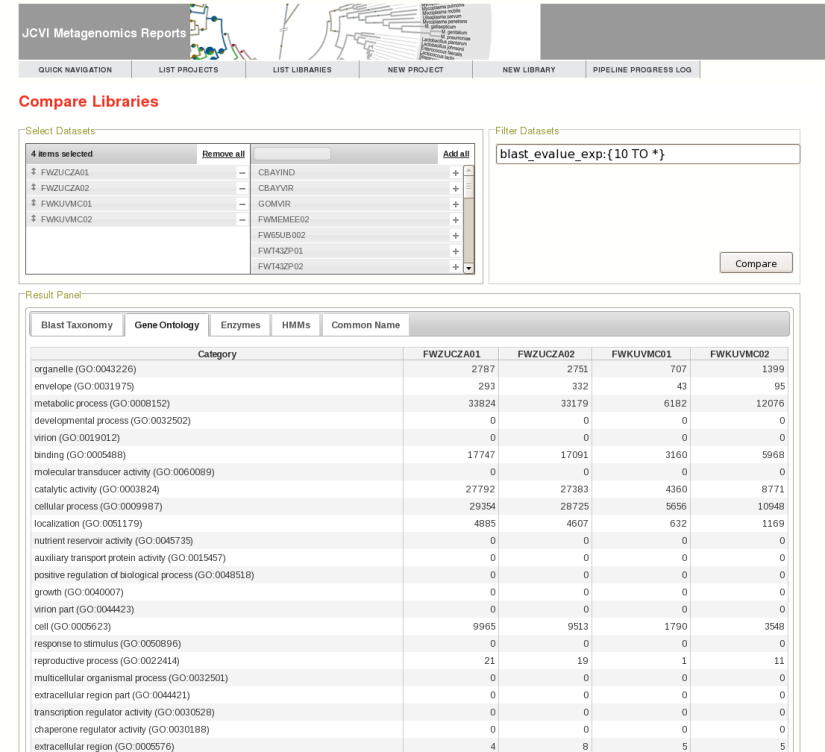
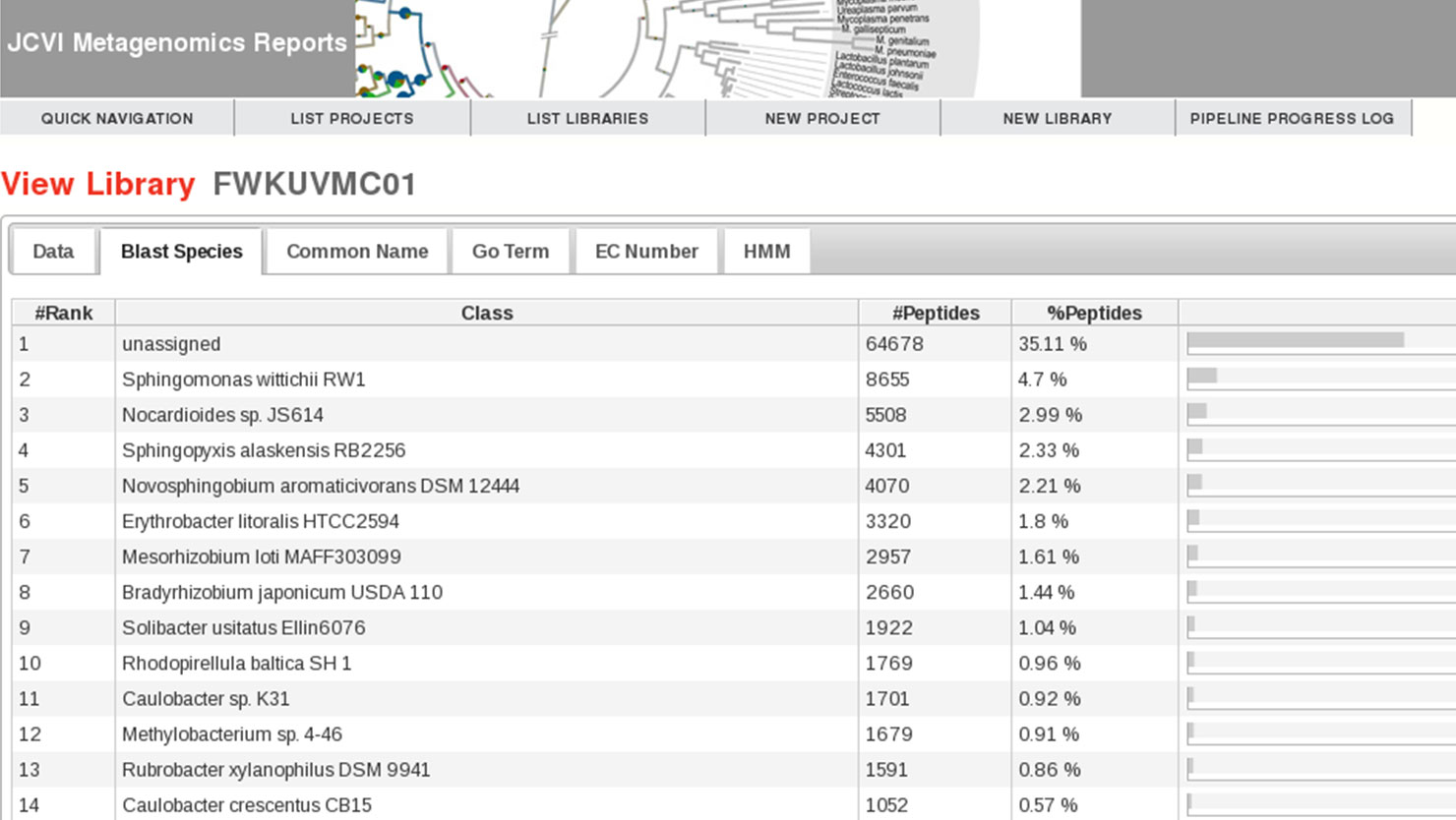
JCVI's Metagenomics Reports (METAREP) is a user friendly web interface designed to help scientists browse, compare, view, and query annotation data derived from ORFs called on metagenomics reads. It supports both functional (Gene Ontology, Enzyme Commission Classification) and browsing of taxonomic assignments. When performing a search, users can either specify fields or logical combinations of fields to flexibly filter datasets on the fly. METAREP provides lists and pie charts of top functional and taxonomic categories for browse and search results. Tools are being developed that focus on the comparative analysis of multiple datasets. The system is optimized to be user friendly and fast .
Currently, an alpha version of METAREP is used and tested internally at JCVI. In April 2010 , we will release the beta version to a limited set of interested external users.
If you like to see the tool in action, join us at the DOE Genomic Science Workshop ( February 9-10, 2010) for our web and poster presentation (5:30 - 8:00 pm on each day) or sign up to become part of the beta testing process at www.jcvi.org/metarep.