Scientists map how iron, a critical mineral for survival, is processed by algae, a cornerstone of the ocean food web
Nearly forty proteins identified in the intracellular process, helping to build a conceptual overview of how iron is allocated within diatom cells
(La Jolla, California)—March 18, 2021—Scientists from the J. Craig Venter Institute (JCVI), Harvard University, and Scripps Institution of Oceanography (SIO) have made significant strides in understanding how iron is processed beneath the cell surface in Phaeodactylum tricornutum, a model marine diatom, publishing their results today in the journal eLife. Diatoms are among the most abundant type of algae found in oceans worldwide contributing up to 40 percent of the 45 to 50 billion metric tons of organic carbon production each year in the sea. Diatoms are able to rapidly consume large amounts of nutrients, fueling some of the most efficient and environmentally vital food webs.
Among these nutrients is iron. Iron likely played an important role in the origin of life and is fundamental to metabolisms of almost all modern life. It is necessary for many key biological processes such as DNA replication, photosynthesis, cellular respiration, nitrate assimilation, nitrogen fixation, and reactive oxygen species defense.
“In vast areas of the modern ocean the growth of phytoplankton, and by extension the ocean carbon cycle, is regulated by the concentration of iron, which can be as scarce as one trillionth of a gram per liter, approaching the limit for supporting life. Therefore, identification of genome-scale adaptations and cellular features that enable survival and proliferation in response to iron limitation and availability are a topic of significant interest in our laboratory,” said Andrew E. Allen, Ph.D., a joint professor between SIO and JCVI, and senior author on the study.
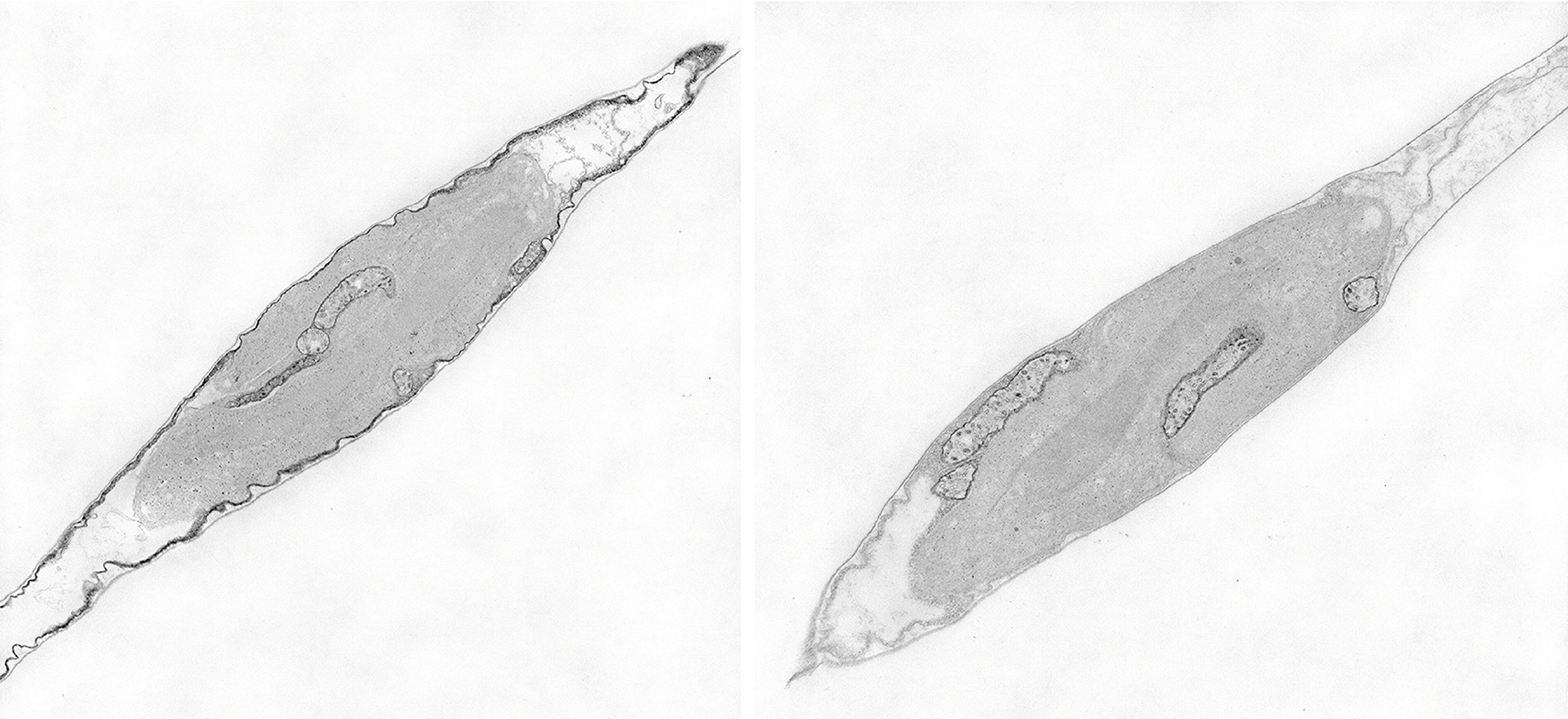
The team previously studied various pathways by which diatoms acquire iron. One of them uses phytotransferrin proteins where iron coordination is contingent on a high carbonate ion concentration in the water. Ocean acidification is reducing carbonate availability making it more difficult for diatoms to collect iron this way. The team discovered another pathway not reliant on carbonate, showing how diatoms may be adapting to changing ocean chemistry by internalizing siderophores, bacterially-produced compounds which coordinate iron.
While these past studies identified and characterized how iron is acquired, nothing was known about how it is trafficked once it passes the cell wall and cell membrane. A proximity proteomics experiment was conducted with APEX2 fused with P. tricornutum phytotransferrin. APEX2 is an emerging proteomics tool which allows for tagging and ultimately cataloguing of proteins near the protein of interest. Almost forty proteins were identified which may constitute the phytotransferrin proximal proteome.
Additional experiments using fluorescence microscopy, phylogenetic, and biochemical analyses, allowed the team to further characterize three proteins that appear to act in association with phytotransferrin, within the cell, downstream of iron binding at the cell surface. The data hints at a possibility for direct cell surface-to-chloroplast iron trafficking pathway, an exciting hypothesis that will require further molecular characterization of the identified proteins. The team synthesized their insights into a new conceptual overview of inorganic iron trafficking in P. tricornutum as a starting point for future projects.
“This work not only adds to the growing molecular tool kit available to study these fascinating single-celled marine eukaryotes, but also opens up questions about evolutionary and molecular parallels of iron acquisition across the tree of life,” said Jernej Turnšek, Ph.D., the lead author of the study who envisioned to apply APEX2 to diatoms as a graduate student at Harvard University. “It is wonderful to see years of efforts spanning both U.S. coasts finally come to fruition,” continued Dr. Turnšek.
Dr. Allen added, “It is clear that our capacity to sequence genomes massively exceeds our ability to decipher the actual function of the many thousands of proteins encoded in these genomes. APEX2 is an exciting method because it represents a powerful means for obtaining robust insights into the function of proteins encoded by poorly understood or completely unknown genes. Jernej’s optimization of APEX2 methodology in diatoms was a perfect fit for our investigations into diatom iron uptake and metabolism and phytotransferrin function. We discovered several proteins that are distributed across a range of diverse marine phytoplankton and appear to be essential for shuttling and directing delivery of iron from phytotransferrin to specific cellular destinations.”
The international research team also included John K. Brunson, a Ph.D. student at the Scripps Institution of Oceanography and J. Craig Venter Institute, Vincent A. Bielinski in the Synthetic Biology and Bioenergy group at the J. Craig Venter Institute, Maria del Pilar Martinez Viedma in the Informatics group at the J. Craig Venter Institute, Thomas J. Deernick from the National Center for Microscopy and Imaging Research at the University of California San Diego, and Miroslav Oborník and Aleš Horák from Biology Centre ASCR, Institute of Parasitology and University of South Bohemia, Czech Republic.
The complete study, Proximity proteomics in a marine diatom reveals a putative cell surface-to-chloroplast iron trafficking pathway, is found in the journal eLife.
About J. Craig Venter Institute
The J. Craig Venter Institute (JCVI) is a not-for-profit research institute in Rockville, Maryland and La Jolla, California. dedicated to the advancement of the science of genomics; the understanding of its implications for society; and communication of those results to the scientific community, the public, and policymakers. Founded by J. Craig Venter, PhD, the JCVI is home to approximately 150 scientists and staff with expertise in human and evolutionary biology, genetics, bioinformatics/informatics, information technology, high-throughput DNA sequencing, genomic and environmental policy research, and public education in science and science policy. The JCVI is a 501(c)(3) organization. For additional information, please visit www.JCVI.org.
Media Contact
Matthew LaPointe, mlapointe@jcvi.org, 301-795-7918